By Elizabeth Hjort, Ph.D. Senior Medical and Scientific Writer at Cognition Studio
Researchers in Seattle have mapped gene expression and DNA accessibility within individual cells across 15 fetal tissues. The findings are presented in back-to-back publications in the November 13, 2020 issue of the journal Science. Together, the studies provide a human cell atlas during development, which may inform our understanding of developmental diseases’ origins.
The gene expression study “is the largest single-cell dataset that has been published up until now. From these data, we can directly generate a catalog of all major cell types across human tissues, including how those cell types might vary in their gene expression across tissues,” said lead author, Junyue Cao. He completed this work as a postdoctoral fellow in the laboratory of Jay Shendure, Professor of Genome Sciences at the University of Washington School of Medicine and Scientific Director of the Brotman Baty Institute for Precision Medicine. Junyue Cao is now an assistant professor at The Rockefeller University.
The map of DNA accessibility uses a new method to understand better how genes are regulated in individual cells. Co-lead author, Silvia Domcke, said, “This new method allows us to profile an order of magnitude more cells than we could previously. This will be a resource for other scientists, and they will be able to use this method for many other types of tissues.” Silvia Domcke is a postdoctoral fellow in the Shendure lab. The other lead authors of the DNA accessibility study are Andrew Hill, previously a graduate student in the Shendure lab and now a scientist at 10x Genomics, and Riza Daza, a research scientist in the Shendure lab.
The human fetal tissues used in these studies were provided by the Birth Defects Research Laboratory (BDRL) at the University of Washington led by Ian Glass, Professor of Pediatrics at the University of Washington. For decades, the BDRL has been a source of donated human fetal tissue for essential medical research. Dan Doherty, Professor of Pediatrics and BDRL co-investigator, explained why the use of human fetal tissues for these studies was important: “This groundbreaking work on human development could not be accomplished in other model systems, because key aspects of human development are unique to humans. While human organoid models have shown promise in recapitulating human development, substantial limitations exist, including limited cellular diversity, lack of mature cell types, and paucity of normal cell-cell interactions.”
In addition to the Brotman Baty Institute and UW Medicine, other collaborators from Illumina, University of Arizona, the Fred Hutchinson Cancer Research Center, the Max Planck Institute for Molecular Genetics and the University of Rochester Medical Center contributed to these studies.
A Human Cell Atlas of Fetal Gene Expression
Gene expression is a process in which a cell uses the information stored in DNA to direct a protein’s production. The activity of the resulting proteins establishes the cell’s structure and function.
In this study, the scientists tracked gene expression within individual cells to understand how it guides human development. “There is an overarching goal of the field to profile the genetic programs that are present in a human at as broad of a scale, and with as much resolution as possible,” said Shendure. “These are emerging as foundational resources for the field. Through the technologies we have developed, we were in a position to take a large step forward towards this goal across a diversity of tissues.”
They profiled gene expression across 15 types of fetal tissue using a technique called sci-RNA-seq3. This technique labels each cell with a unique combination of three DNA “barcodes”, allowing the researchers to keep track of cells without physically separating them. Once the sequences were obtained, they used computer algorithms to recover the single-cell information, cluster cells by type and subtype, and identify developmental trajectories.
The scientists profiled over 4 million single cells, identified 77 main cell types and approximately 650 cell subtypes. The data’s breadth and depth allowed them to characterize the dynamics of gene expression across multiple organs, not just within one organ. For example, they discovered more about how the endothelial cells that line blood vessels differ from one another across multiple organs.
They detected some extremely rare cell types, and they discovered that red blood cells are made in the adrenal gland during human development.
They also integrated this human single cell atlas with a single cell atlas of mouse embryonic development that they had previously released. Co-senior author Cole Trapnell, associate professor of genome sciences at the University of Washington School of Medicine, explained, “When we combine this data with previously published data, we can directly delineate cells’ developmental path for all major cell types.”
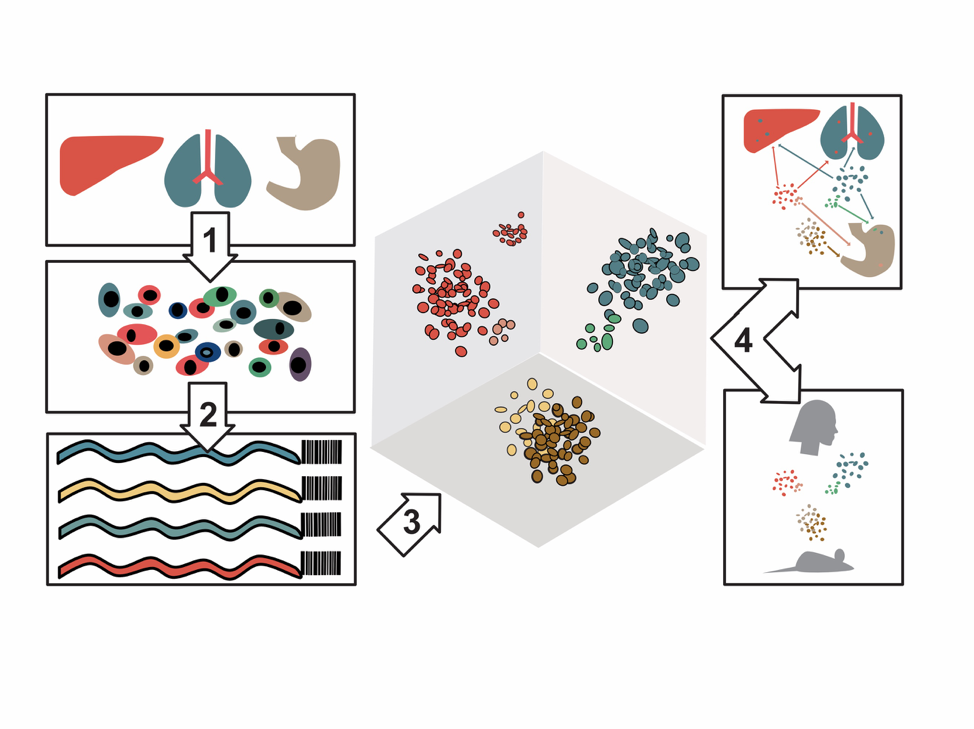 Gene expression and chromatin accessibility profiles of individual cells from 15 different organs were created in the study. Image by Michael McCarthy.
Gene expression and chromatin accessibility profiles of individual cells from 15 different organs were created in the study. Image by Michael McCarthy.
A Human Cell Atlas of Fetal Chromatin Accessibility
The three-dimensional structure that allows DNA to pack tightly into the nucleus of the cell is called chromatin. Chromatin can be open and “accessible”, or closed and “inaccessible”. Knowing the patterns and regions of DNA that are open versus regions that are closed can tell us more about how the cell chooses to turn genes on and off, both in healthy cells and in disease.
Previously, there have been very few resources to distinguish DNA accessibility patterns at the single-cell level. Researchers need to be able to understand these patterns because they are related to the genetic factors that influence our risk for common human diseases. For example, cancer, autoimmune diseases and neurological diseases are all impacted by DNA accessibility.
“Up until now, most of the focus in the field has been on single-cell atlases of gene expression. Studying chromatin gives you a sense of the regulatory “grammar” of the cell,” co-senior author Darren Cusanovich, previously a postdoctoral fellow in the Shendure lab and now an assistant professor at the University of Arizona, said. “The short stretches of DNA that are open, or accessible, are enriched for certain “words,” which are, in turn, the basis for the cell to specify that it wants certain genes on.”
To profile DNA accessibility in individual cells, the scientists invented a new method, called sci-ATAC-seq3. Like sci-RNA-seq3, this technique also uses three different DNA “barcodes” in each cell to tag and track individual cells. However, rather than identifying all the currently expressed sequences, sci-ATAC-seq3 captures and sequences open chromatin sites.
The scientists generated nearly 800,000 single-cell chromatin accessibility profiles at approximately 1 million sites across 15 fetal organs in this study. They asked which proteins were likely to interact with accessible DNA sites in each cell and how those interactions can explain the cell type. This analysis defines the control switches for development within the genome. They also identified sites of chromatin accessibility that might be associated with diseases.
Domcke explained, “This study tells us what part of the genome might be functional. We still do not know what percentage of the genome that doesn’t encode for genes can be involved in gene regulation. Our atlas now provides that information for many cell types.”
Together, these studies provide a fundamental resource for understanding gene expression and chromatin accessibility in human development at an unprecedented scale. The techniques that are described in these publications can be used in tandem to generate data on gene expression and chromatin accessibility for millions of cells.
These scientists are paving the way to expand single-cell sequencing at other time points in development and establish a more predictive investigation of gene regulation in human diseases.
Learn more in the Science publications, “A Human Cell Atlas of Fetal Gene Expression,” and “A Human Cell Atlas of Chromatin Accessibility.” These data can be interactively visualized at a website developed by Michael Zager at the Fred Hutchinson Cancer Research Center and Brotman Baty Institute Center for Data Visualization at https://descartes.brotmanbaty.org/
Different components of these studies were funded by the Brotman Baty Institute for Precision Medicine, the Paul G. Allen Frontiers Foundation, the Chan Zuckerberg Initiative, Howard Hughes Medical Institute, and the National Institutes of Health.


